This vignette covers topics related to alpha diversity and endemism, including calculation of basic diversity metrics as well as statistical significance testing using null randomization.
To get started, let’s load the phylospatial library, as
well as tmap for visualization. Note that the functions
covered here all require a phylospatial object as input;
see vignette("phylospatial-data") for details on
constructing data sets. We’ll use the moss() example data
here.
library(phylospatial); library(tmap)
ps <- moss()Diversity measures
The ps_diversity() function calculates the following
alpha diversity measures. While there are a wide variety of
phylogentically-informed diversity metrics in the literature, the
phylospatial package focuses primarily on “node-based”
diversity metrics like Faith’s PD that give equal treatment to
clades/branches at all levels. Phylogenetic diversity metrics can be
categorized as addressing richness, divergence, and regularity (Tucker
et al. 2017), as listed below. Note that an alternative approach to
assessing divergence is through null model analysis of richness metrics,
discussed later in this vignette.
Richness metrics:
- PD: Phylogenetic diversity, i.e. total length of all branch segments present in a location
- PE: Phylogenetic endemism, i.e. endemism-weighted PD
- ShPD: Shannon’s pylogenetic diversity, a.k.a “phylogenetic entropy”
- ShPE: Shannon’s pylogenetic diversity, weighted by endemism
- SiPD: Simpson’s phylogenetic diversity
- SiPE: Simpson’s phylogenetic diversity, weighted by endemism
- TR: Terminal richness, i.e. richness of terminal taxa (in many cases these are species). For binary data this is the total number of terminals in a site, while for quantitative data it is the sum of probability or abundance values.
- TE: Terminal endemism, i.e. total endemism-weighted diversity of terminal taxa (a.k.a. “weighted endemism”)
- CR: Clade richness, i.e. richness of taxa at all levels (equivalent to PD on a cladogram)
- CE: Clade endemism, i.e. total endemism-weighted diversity of taxa at all levels (equivalent to PE on a cladrogram)
Divergence metrics:
- RPD: Relative phylogenetic diversity, i.e. mean branch segment length of residents (equivalent to PD / CR)
- RPE: Relative phylogenetic endemism, i.e. mean endemism-weighted branch length (equivalent to PE / CE)
- MPDT: Mean pairwise distance between terminals, i.e. the classic MPD measure
- MPDN: Mean pairwise distance between nodes, a node-based version of MPD that calculates the average branch length separating all pairs of collateral (non-lineal) nodes including terminals and internal nodes, giving more representation to deeper branches
Regularity metrics:
- VPDT: Variance in pairwise distances between terminals
- VPDN: Variance in pairwise distances between collateral nodes
All measures use quantitative community data if provided. “Endemism”
is the inverse of the total occurrence mass (the sum of presence,
probability, or abundance values) across all sites in the analysis. See
?ps_diversity for equations giving the derivation of each
metric.
Let’s compute some diversity metrics for our phylospatial data set.
Since our data is raster-based, by default the function will return a
SpatRaster with a layer for each measure. Here we’ll make
plots of PD and PE:
div <- ps_diversity(ps, metric = c("PD", "PE"))
tm_shape(div$PD) +
tm_raster(col.scale = tm_scale_continuous(values = "inferno")) +
tm_layout(legend.outside = TRUE)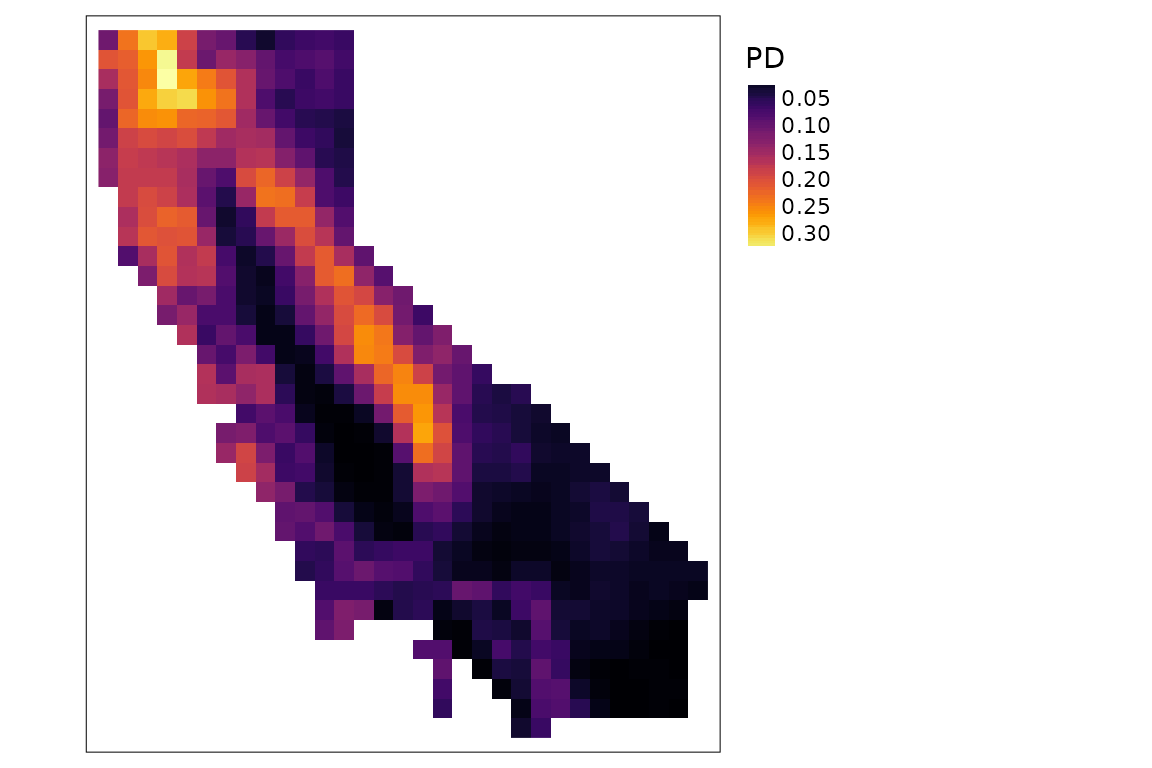
tm_shape(div$PE) +
tm_raster(col.scale = tm_scale_continuous(values = "inferno")) +
tm_layout(legend.outside = TRUE)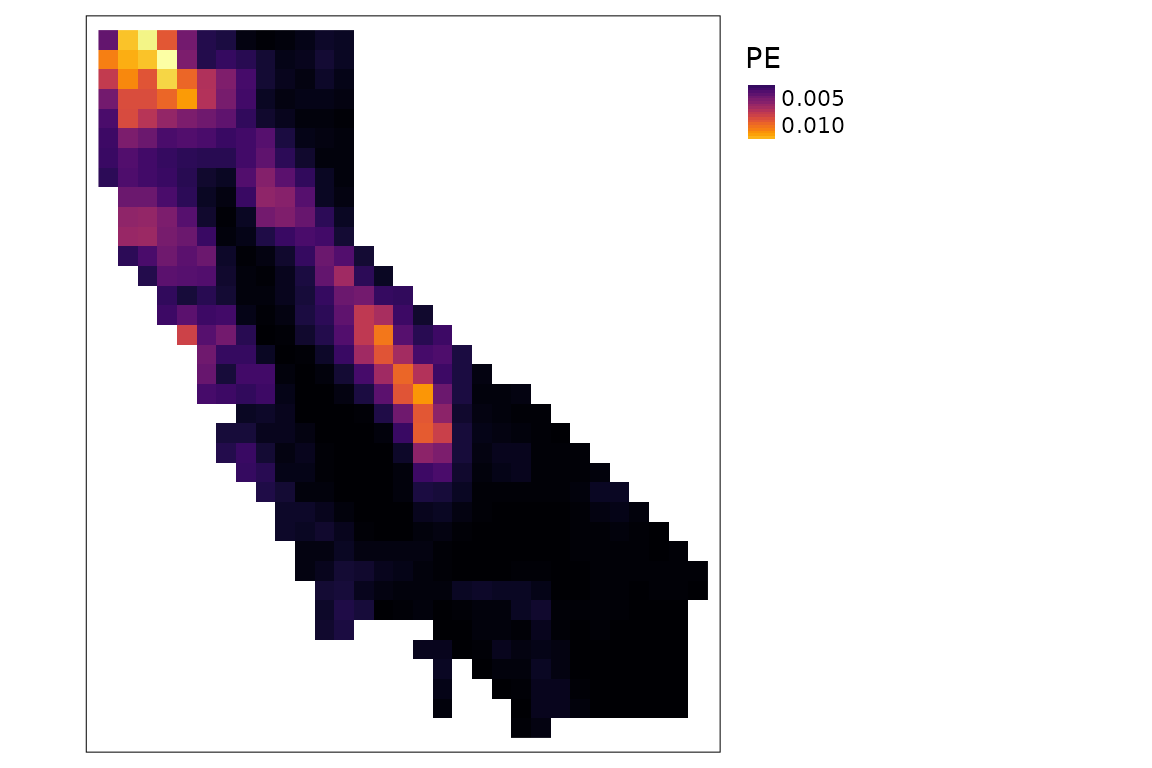
Null model randomization
We can also use randomization to calculate the statistical
significance of these diversity metrics under a null model, using the
ps_rand() function. Here let’s run our randomization using
quantize, a stratified randomization scheme designed for
use with continuous occurrence data, in combination with a null model
algorithm called "curvecat", which is a categorical version
of the “curveball” algorithm that holds marginal row and column
multisets fixed. (Note that this categorical null model requires the
nullcat library.)
We’ll run 1000 randomizations for four diversity metrics, and plot the results for PE. This is a quantile value that gives the proportion of randomizations in which observed PE was greater than randomized PE in a given grid cell. (If you wanted to identify “statistically significant” grid cells in a one-tailed test with alpha = 0.05, these would be cells with values greater than 0.95.)
rand <- ps_rand(ps, n_rand = 1000, progress = FALSE,
metric = c("PD", "PE", "CE", "RPE"),
fun = "quantize", method = "curvecat")
tm_shape(rand$qPE) +
tm_raster(col.scale = tm_scale_continuous(values = "inferno")) +
tm_layout(legend.outside = TRUE)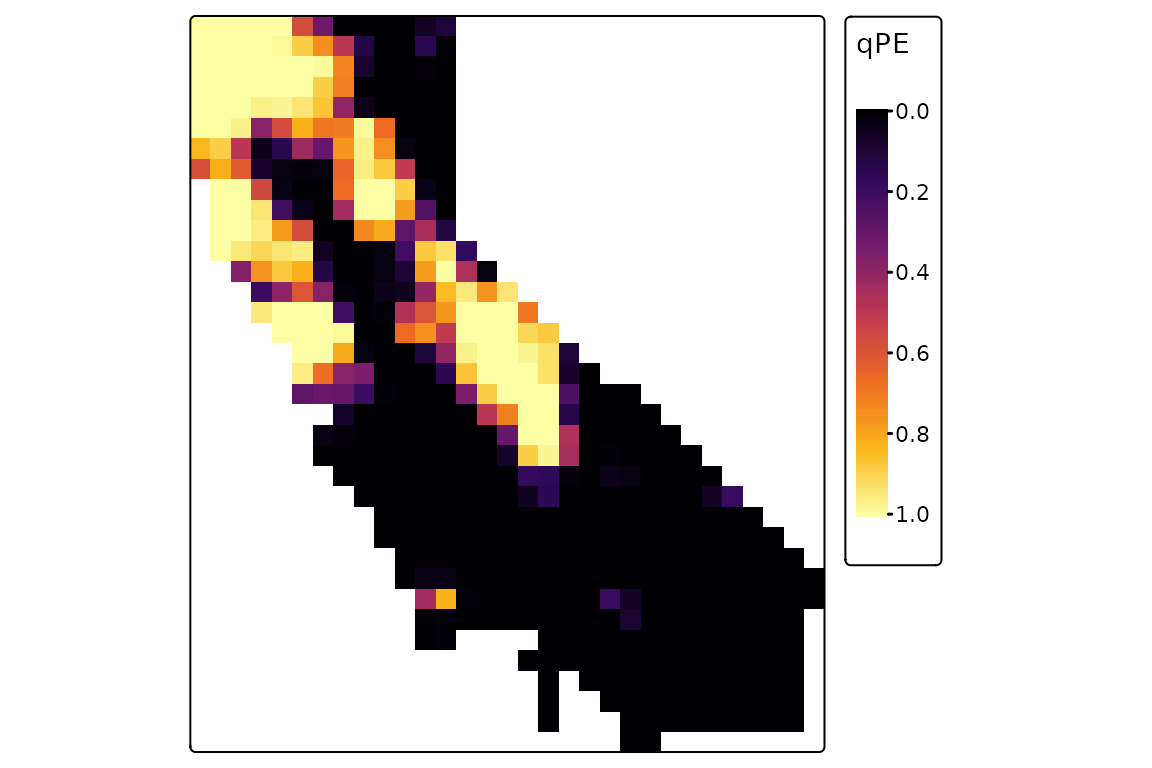
There are numerous alternative options for randomization algorithms,
a choice that will depend on the type of occurrence data you have
(probability, binary, or abundance) and on which attributes of the
terminal community matrix (fill, row and column sums, etc.) you want to
hold fixed. In addition to the quantize function used
above, these include a basic "tip_shuffle" randomization
(the default algorithm), a range of algorithms defined in the
vegan package, and an option to supply a custom
randomization function. As a second example, here’s a randomization with
an abundance data set, using the "abuswap_c" algorithm
provided by vegan::nullmodel:
ps2 <- ps_simulate(data_type = "abundance")
rand2 <- ps_rand(ps2, fun = "nullmodel", method = "abuswap_c", progress = FALSE, metric = "PD")CANAPE
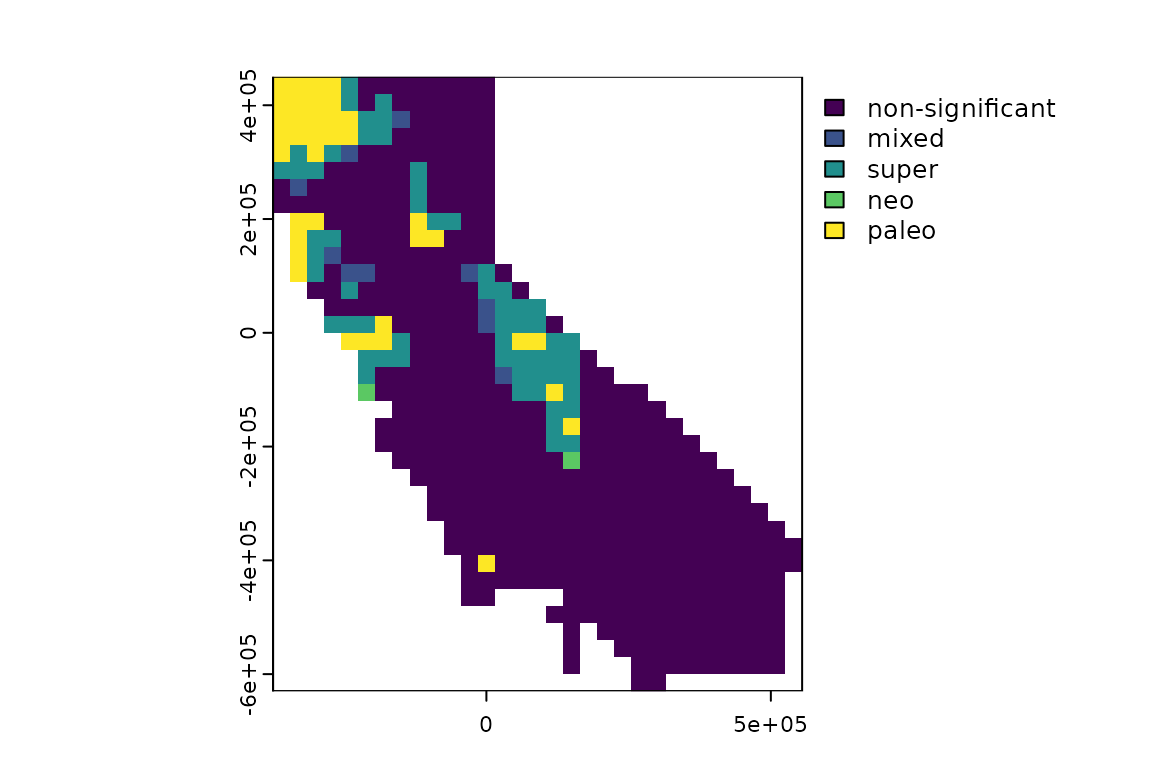
Many things can be done with randomization results like the ones we
generated above. One thing you can do is use them to classify
significant endemism hotspots in a “categorical analysis of neo- and
paleo-endemism” (CANAPE, Mishler et al. 2014). The
function ps_canape() uses significance values for PE, RPE,
and CE, which are returned by ps_rand(), to categorize
sites into five endemism cateogories. Here’s an example with the moss
data; note that depending on the randomization, only a subset of the
five categories may occur in the result here: