Introduction
nullcat provides null model algorithms for categorical
and quantitative community ecology data. It extends classic binary null
models (e.g., curveball, swap) to work with categorical data, and
introduces a stratified randomization framework for continuous data.
Categorical null models generalize binary null models to matrices where cells contain integers representing discrete categories. Whereas binary models preserve totals (e.g. row or column sums), categorical models preserve multisets (e.g. category frequencies in each row or column). For example, if a row contains [1, 1, 2, 3, 3, 3], a row-preserving categorical algorithm would preserve this exact multiset—two 1s, one 2, and three 3s—while randomizing where these categories appear within the row.
Traditional binary models are simply the specific two-category cases
of their corresponding general categorical models
(e.g. curveball is equivalent to curvecat with
only two categories).
Use cases
Categorical null models are designed for cell-level categorical data—characteristics of a species at a site—rather than site-level or species-level attributes. They can be useful for categorical and ordinal ecological data. Examples include:
- Ordinal states: phenological stages, health status, life stages
- Multi-state interactions: interaction type, infection status, mutualism outcomes
- Population genetic states: genotypes or allelic frequencies at the population level
- Binned quantitative measures: abundance classes, cover scales, or
occurrence probability bins (the
quantize()function covered below automates binning and randomization) - Binary data: species presence-absence data or similar
Categorical null models
Available algorithms
The package provides five categorical null model algorithms:
-
curvecat(): Categorical curveball (most efficient) -
swapcat(): Categorical 2×2 swap -
tswapcat(): Categorical trial-swap -
r0cat(): Row-constrained randomization -
c0cat(): Column-constrained randomization
Example: Basic categorical randomization
# Create a categorical matrix
set.seed(123)
x <- matrix(sample(1:4, 100, replace = TRUE), nrow = 10)
# Randomize using curvecat (preserves row & column category multisets)
x_rand <- curvecat(x, n_iter = 1000)
# Verify margins are preserved
all.equal(sort(x[1, ]), sort(x_rand[1, ])) # Row preserved
#> [1] TRUE
all.equal(sort(x[, 1]), sort(x_rand[, 1])) # Column preserved
#> [1] TRUEUnderstanding fixed margins
Different algorithms preserve different margins:
set.seed(456)
x <- matrix(sample(1:3, 60, replace = TRUE), nrow = 10)
# Fixed-fixed: both row and column margins preserved
x_fixed <- curvecat(x, n_iter = 1000)
# Row-constrained: only row margins preserved
x_row <- r0cat(x)
# Column-constrained: only column margins preserved
x_col <- c0cat(x)
# Check row sums by category
table(x[1, ]) # Original row 1
#>
#> 1 2 3
#> 2 2 2
table(x_fixed[1, ]) # Preserved in curvecat
#>
#> 1 2 3
#> 2 2 2
table(x_row[1, ]) # Preserved in r0cat
#>
#> 1 2 3
#> 2 2 2
table(x_col[1, ]) # Not preserved in c0cat
#>
#> 1 2 3
#> 1 3 2Quantitative null models
The quantize() function provides stratified
randomization for continuous community data (abundance, biomass, cover,
occurrence probability). It works by applying the following steps:
- Stratify: use a flexible binning scheme to convert continuous values into discrete strata
- Randomize: apply one of the categorical null models described above to the stratified matrix
- Reassign: map randomized strata back to original values, subject to a chosen constraint
Why use quantize? Existing quantitative null models
lack flexibility in what margins they preserve. While binary curveball
elegantly preserves row and column totals, and vegan’s quasiswap models
do this for count data, there’s no framework for preserving row and/or
column value distributions for continuous data
(abundance, biomass, cover). The stratification approach fills this gap:
when using fixed = "row" or fixed = "col", one
dimension’s value multisets are preserved exactly while the other is
preserved at the resolution of strata. This is the closest available
approximation to a quantitative fixed-fixed null model. Additionally,
stratification provides explicit control over the
preservation-randomization tradeoff through the number of strata
used.
Example: Basic quantitative randomization
# Create a quantitative community matrix
set.seed(789)
comm <- matrix(runif(200), nrow = 20)
# Default: curvecat-backed stratified randomization with 5 strata
rand1 <- quantize(comm, n_strata = 5, n_iter = 2000, fixed = "row")
# Values are similar but rearranged
cor(as.vector(comm), as.vector(rand1))
#> [1] 0.1757922
plot(rowSums(comm), rowSums(rand1))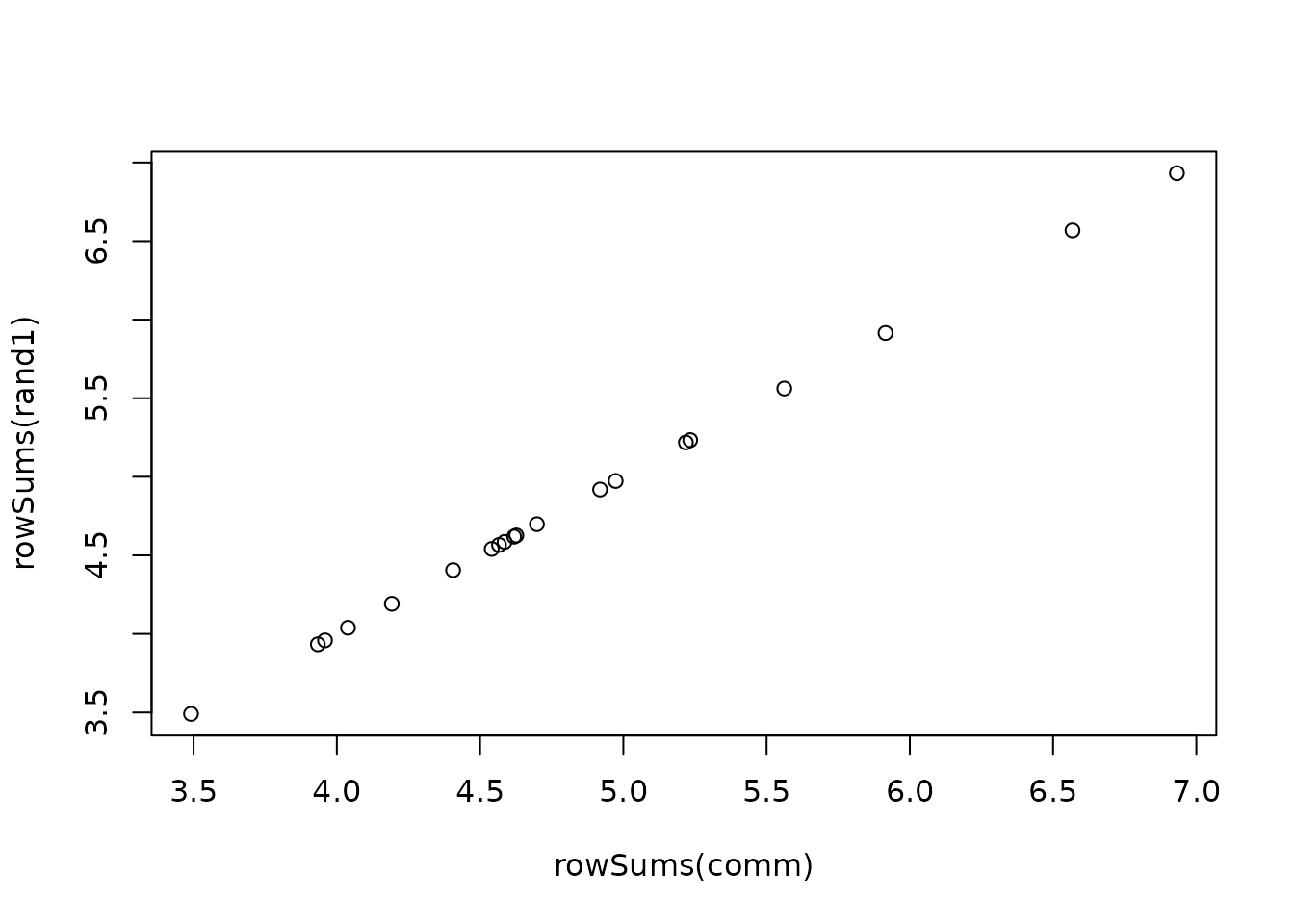
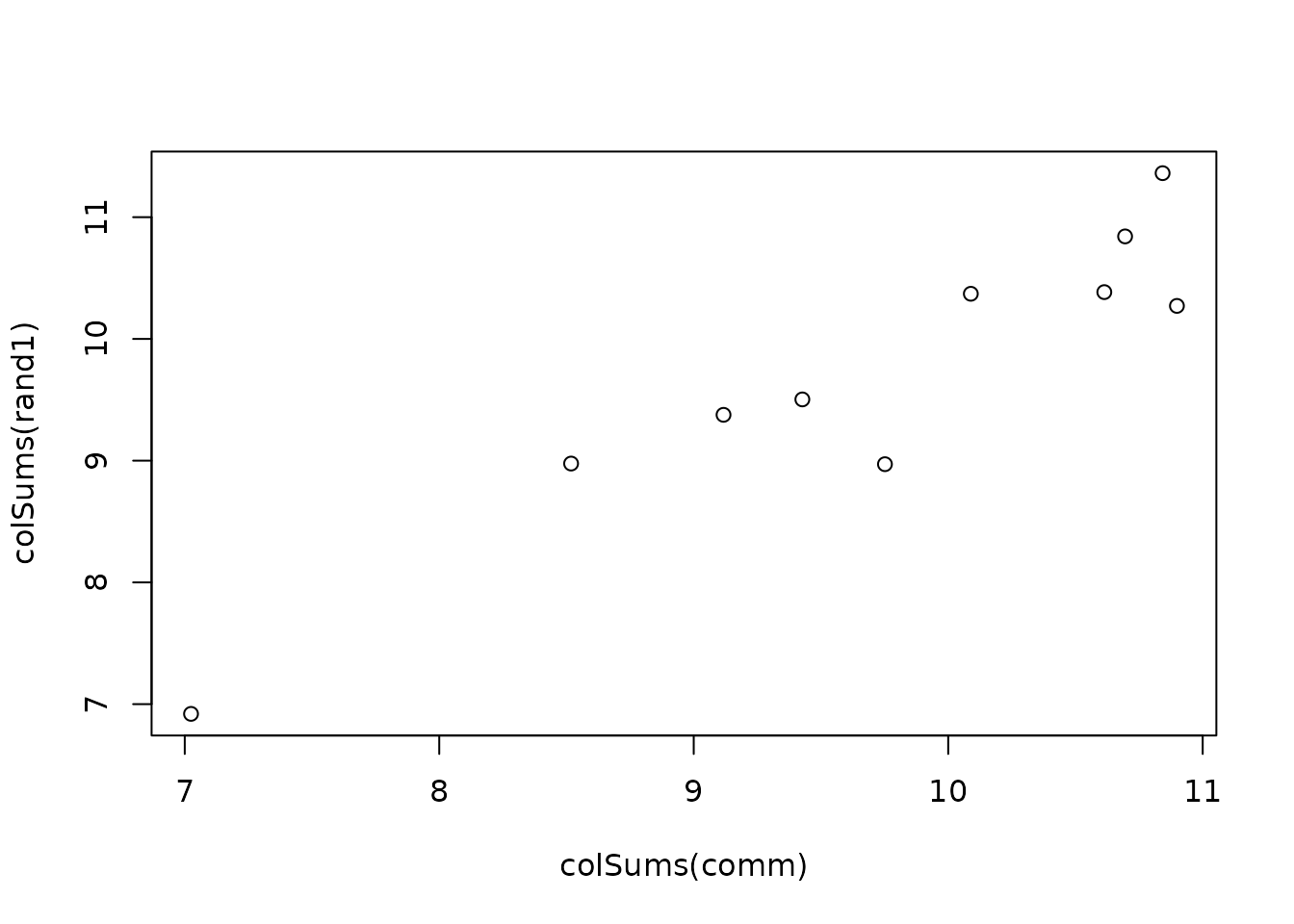
Stratification options
When you call quantize(), it uses the helper function
stratify() to convert your continuous data to discrete
bins. The stratification scheme can strongly influence your
randomization results, because it is frequencies of these categories
that are constrained during matrix permutation. By default, data are
split into a handful of equal-width bins, but this can be controlled
with parameters including n_strata, transform,
zero_stratum, breaks, and offset.
Let’s call stratify() directly here, to understand its
behavior—a good idea for a real analysis, not just a tutorial.
set.seed(200)
x <- rexp(100, .1)
# More strata = less mixing, higher fidelity to original distribution
s3 <- stratify(x, n_strata = 3)
s10 <- stratify(x, n_strata = 10)
table(s3) # Coarser bins
#> s3
#> 1 2 3
#> 89 9 2
table(s10) # Finer bins
#> s10
#> 1 2 3 4 5 6 10
#> 49 22 17 4 4 2 2
# Transform before stratifying (e.g., log-transform for skewed data)
s_log <- stratify(x, n_strata = 5, transform = log1p)
table(s_log)
#> s_log
#> 1 2 3 4 5
#> 14 26 27 27 6
# Rank transform creates equal-occupancy strata
s_rank <- stratify(x, n_strata = 5, transform = rank)
table(s_rank) # Nearly equal counts per stratum
#> s_rank
#> 1 2 3 4 5
#> 20 20 20 20 20
# Separate zeros into their own stratum
x_with_zeros <- c(0, 0, 0, x)
s_zero <- stratify(x_with_zeros, n_strata = 4, zero_stratum = TRUE)
table(s_zero)
#> s_zero
#> 1 2 3 4
#> 3 89 9 2Controlling what’s preserved
While the mechanics of the chosen randomization algorithm determine
which margins of the randomized categories are preserved, the
fixed parameter controls how the original
continuous values are mapped back onto the randomized
categories, determining which margins preserve their original value
distributions. There are four options for the fixed
parameter: row, col, cell, and stratum, though only a subset of these
are relevant for some algorithms. For a method like the default
curvecat algorithm that preserves both row and column
categorical multisets, all four are available:
set.seed(100)
comm <- matrix(rexp(100), nrow = 10)
# Preserve row value multisets (quantitative row sums maintained)
rand_row <- quantize(comm, n_strata = 5, fixed = "row", n_iter = 2000)
all.equal(rowSums(comm), rowSums(rand_row))
#> [1] TRUE
# Preserve column value multisets (quantitative column sums maintained)
rand_col <- quantize(comm, n_strata = 5, fixed = "col", n_iter = 2000)
all.equal(colSums(comm), colSums(rand_col))
#> [1] TRUE
# Cell-level preservation: each value moves with its original cell location
# The categorical randomization determines WHERE cells go, but each cell
# carries its original value with it
rand_cell <- quantize(comm, n_strata = 5, fixed = "cell", n_iter = 2000)
# Values shuffled globally within strata, holding none of the above fixed
rand <- quantize(comm, n_strata = 5, fixed = "stratum", n_iter = 2000)
# For non-fixed-fixed methods like r0cat or c0cat, only some fixed options make sense:
# r0cat with fixed="col" or c0cat with fixed="row" would be incompatibleEfficient repeated randomization
The quantize routine includes some setup, including stratification,
that doesn’t need to be repeatedly recomputed when generating multiple
randomized versions of the same matrix. For generating null
distributions, you can pre-compute overhead once using
quantize_prep(), or you can use *_batch()
functions to efficiently generate multiple replicates (optionally using
parallel processing).
set.seed(400)
comm <- matrix(rexp(200), nrow = 20)
# Prepare once
prep <- quantize_prep(comm, n_strata = 5, fixed = "row", n_iter = 2000)
# Generate many randomizations efficiently
rand1 <- quantize(prep = prep)
rand2 <- quantize(prep = prep)
rand3 <- quantize(prep = prep)
# Or use batch functions
nulls <- quantize_batch(comm, n_reps = 99, n_strata = 5,
fixed = "row", n_iter = 2000)
dim(nulls) # 20 rows × 10 cols × 99 replicates
#> [1] 20 10 99Convergence diagnostics
Sequential algorithms (curvecat, swapcat, tswapcat) require
sufficient iterations to reach stationarity. You can use the
trace_cat() function with either nullcat or
quantize to assess how many iterations are needed for your
particular dataset and algorithm. The trace shows how similarity to the
original matrix changes over iterations; when the trace stabilizes
around zero, the algorithm has reached stationarity. You can also use
suggest_n_iter() to get an automated suggestion for a
reasonable number of burn-in iterations to reach stationarity.
set.seed(300)
x <- matrix(sample(1:5, 400, replace = TRUE), nrow = 20)
# Generate trace showing mixing over iterations
trace <- trace_cat(x, fun = "nullcat", method = "curvecat",
n_iter = 1000, n_chains = 3, thin = 10)
# Visual inspection
plot(trace)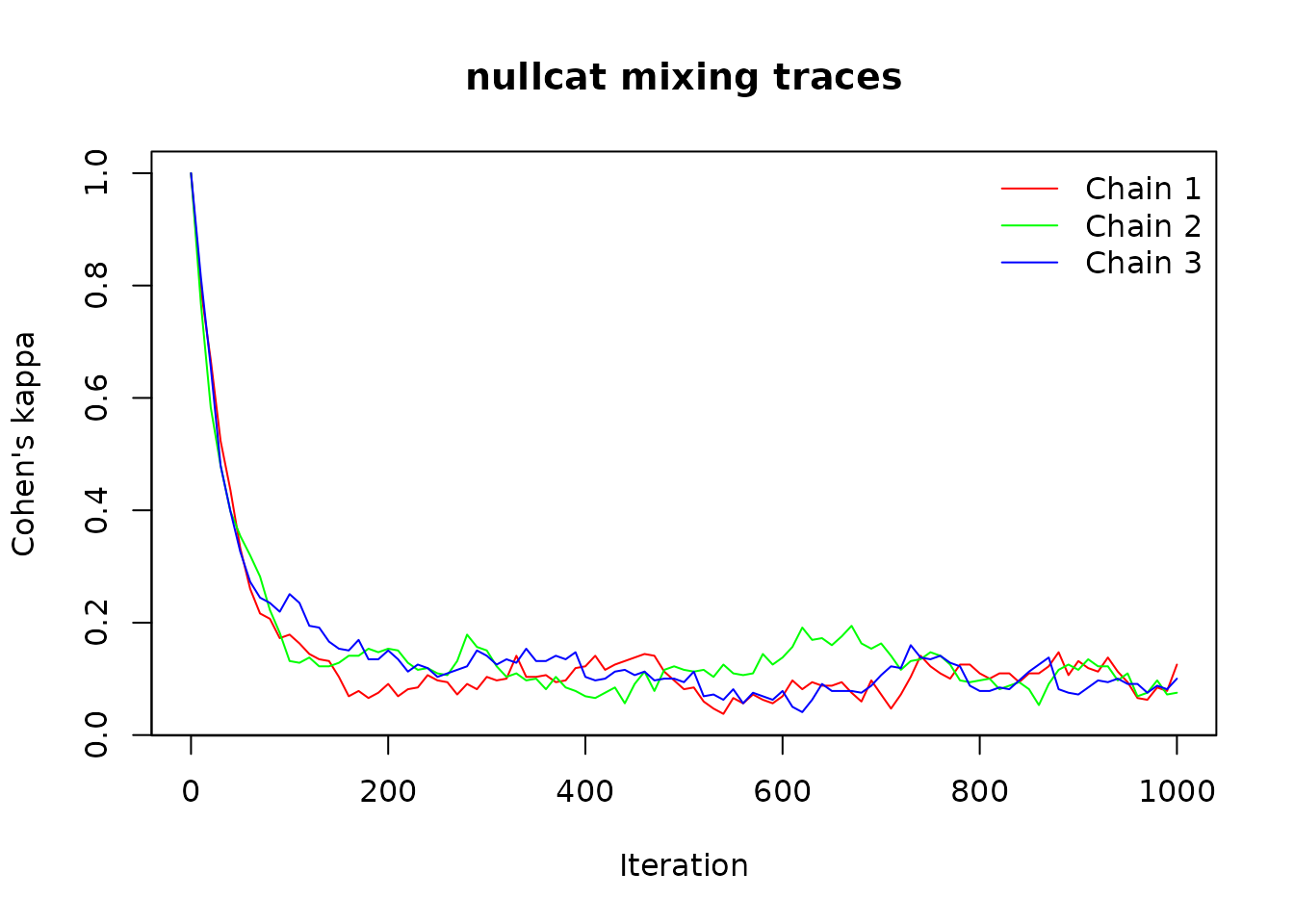
# Automatic burn-in suggestion
suggested <- suggest_n_iter(trace, tail_frac = 0.3)
print(suggested)
#> suggested_n_iter object
#> -----------------------
#> Converged: TRUE
#> Suggested n iterations: 410Integration with vegan
nullcat includes several functions to integrate with the
vegan package’s null model framework. For both
nullcat() and quantize(), there are sequential
and non-sequential variants of these functions that create
vegan::commsim() objects suitable for use with
vegan::nullmodel() and vegan::oecosimu().
Examples include:
library(vegan)
# Categorical data
x_cat <- matrix(sample(1:4, 100, replace = TRUE), nrow = 10)
cs <- nullcat_commsim_seq(method = "curvecat")
nm <- nullmodel(x_cat, cs)
sims <- simulate(nm, nsim = 99, burnin = 1000, thin = 100)
# Quantitative data
x_quant <- matrix(rexp(100), nrow = 10)
cs_quant <- quantize_commsim(n_strata = 5, method = "curvecat", n_iter = 2000)
nm_quant <- nullmodel(x_quant, cs_quant)
sims_quant <- simulate(nm_quant, nsim = 99)References
- Strona et al. (2014). A fast and unbiased procedure to randomize ecological binary matrices. Nature Communications, 5, 4114.
- Gotelli (2000). Null model analysis of species co-occurrence patterns. Ecology, 81(9), 2606-2621.
Further reading
- Package website: https://matthewkling.github.io/nullcat/
- Report issues: https://github.com/matthewkling/nullcat/issues
