phylospatial
[PUBLICATION; R PACKAGE] A toolkit for spatial phylogenetic analysis.
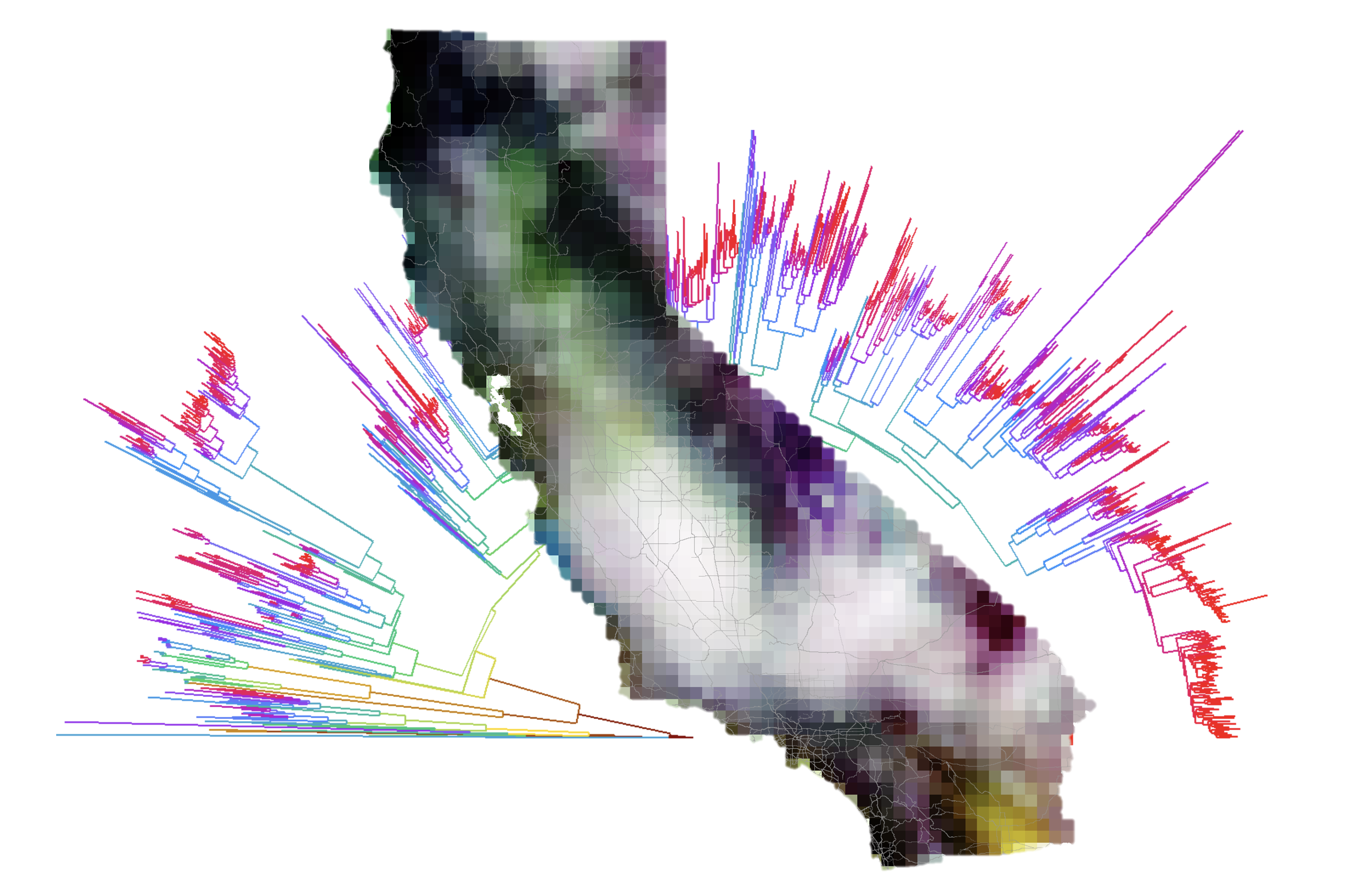
Overview
phylospatial is an R package for spatial phylogenetic analysis, a field that focuses on accounting for evolutionary relationships among taxa when describing biodiversity patterns. The package grew out of code I developed for research on plant diversity and conservation in California, and is described in a paper in Methods in Ecology and Evolution. It provides a flexible framework for analyzing biodiversity patterns that incorporates both geographic and evolutionary dimensions.
Key Features
- Core phylospatial data structure that integrates occurrence data, phylogenetic trees, and spatial context
- Alpha diversity metrics including phylogenetic diversity, endemism, relative phylogenetic diversity, and Shannon/Simpson indices
- Statistical significance testing using null model randomizations with specialized algorithms for quantitative data
- Beta diversity analysis with phylogenetic dissimilarity measures for turnover and nestedness
- Visualization tools including phylogenetic regionalization and ordination
- Conservation prioritization with algorithms for optimal or probabilistic protected area selection
A key innovation is that unlike other spatial phylogenetic libraries, all functions in this package work with continuous community data (occurrence probabilities or abundances) in addition to binary presence-absence data. These quantities are treated as weights in all computations, avoiding the need to discard information by thresholding continuous data.
Applications
The package is particularly useful for:
- Identifying evolutionary “hotspots” for conservation prioritization
- Examining spatial patterns of phylogenetic composition
- Testing hypotheses about the processes that shape biodiversity patterns
- Creating conservation plans that explicitly value evolutionary history
- Analyzing biogeographic patterns with a phylogenetic perspective
Resources
Citation: Kling, M.M. (2025). phylospatial: an R package for spatial phylogenetic analysis with quantitative community data. Methods in Ecology and Evolution, 2025;00:1–9 https://doi.org/10.1111/2041-210X.70056
The package is available on CRAN and includes a comprehensive set of vignettes covering data handling, alpha diversity, beta diversity, and conservation prioritization workflows. The development version is available on GitHub, where you can also find detailed documentation and examples.
